What Is The Most Recent Common Ancestor Of Fungi And Animals Quizlet
In biology and genetic genealogy, the most contempo common antecedent (MRCA), besides known every bit the last common antecedent (LCA) or concestor,[note i] of a set of organisms is the near recent individual from which all the organisms of the set are descended. The term is also used in reference to the beginnings of groups of genes (haplotypes) rather than organisms.
The MRCA of a set of individuals can sometimes be determined by referring to an established pedigree. However, in general, it is incommunicable to identify the exact MRCA of a large set up of individuals, but an guess of the time at which the MRCA lived can often be given. Such time to most recent common ancestor (TMRCA) estimates can be given based on DNA exam results and established mutation rates as practiced in genetic genealogy, or by reference to a non-genetic, mathematical model or computer simulation.
In organisms using sexual reproduction, the matrilineal MRCA and patrilineal MRCA are the MRCAs of a given population considering just matrilineal and patrilineal descent, respectively. The MRCA of a population by definition cannot exist older than either its matrilineal or its patrilineal MRCA. In the instance of Human sapiens, the matrilineal and patrilineal MRCA are also known every bit "Mitochondrial Eve" (mt-MRCA) and "Y-chromosomal Adam" (Y-MRCA) respectively.
The age of the human MRCA is unknown. It is no greater than the age of either the Y-MRCA or the mt-MRCA, estimated at effectually 200,000 years.
Different in pedigrees of individual humans or domesticated lineages where historical parentage is known, in the inference of relationships amongst species or higher groups of taxa (systematics or phylogenetics), ancestors are not directly appreciable or recognizable. They are inferences based on patterns of human relationship amid taxa inferred in a phylogenetic assay of extant organisms and/or fossils. [1]
The terminal universal common ancestor (LUCA) is the about recent common ancestor of all current life on Earth, estimated to take lived some 3.5 to 3.viii billion years agone (in the Paleoarchean).[2] [3] [note 2]
MRCA of different species [edit]
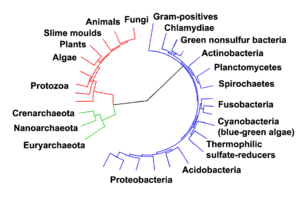
The project of a complete description of the phylogenetic relationships amidst all biological species is dubbed the "tree of life". This involves inference of ages of divergence for all hypothesized clades; for case, the MRCA of all Carnivora (i.eastward. the MRCA of "cats and dogs") is estimated to have diverged some 42 million years ago (Miacidae).[6]
The concept of the last common antecedent from the perspective of human development is described for a popular audition in The Ancestor'southward Tale by Richard Dawkins (2004). Dawkins lists "concestors" of the human lineage in order of increasing historic period, including hominin (man–chimpanzee), hominine (human–gorilla), hominid (man–orangutan), hominoid (human–gibbon), then on in xl stages in total, down to the terminal universal ancestor (human being–bacteria).
MRCA of a population identified by a single genetic marker [edit]
It is likewise possible to consider the ancestry of individual genes (or groups of genes, haplotypes) instead of an organism as a whole. Coalescent theory describes a stochastic model of how the ancestry of such genetic markers maps to the history of a population.
Unlike organisms, a factor is passed down from a generation of organisms to the next generation either as perfect replicas of itself or as slightly mutated descendant genes. While organisms have beginnings graphs and progeny graphs via sexual reproduction, a gene has a unmarried chain of ancestors and a tree of descendants. An organism produced past sexual cross-fertilization (allogamy) has at least two ancestors (its firsthand parents), but a gene e'er has one ancestor per generation.
Patrilineal and matrilineal MRCA [edit]

Through random drift or pick, lineage will trace back to a single person. In this case over five generations, the colors represent extinct matrilineal lines and black the matrilineal line descended from the mt-MRCA.
Mitochondrial Deoxyribonucleic acid (mtDNA) is virtually immune to sexual mixing, unlike the nuclear DNA whose chromosomes are shuffled and recombined in Mendelian inheritance. Mitochondrial DNA, therefore, can exist used to trace matrilineal inheritance and to observe the Mitochondrial Eve (also known as the African Eve), the most recent common ancestor of all humans via the mitochondrial DNA pathway.
Also, Y chromosome is present as a single sexual practice chromosome in the male person private and is passed on to male person descendants without recombination. It can be used to trace patrilineal inheritance and to discover the Y-chromosomal Adam, the near contempo common ancestor of all humans via the Y-Deoxyribonucleic acid pathway.
Approximate dates for Mitochondrial Eve and Y-chromosomal Adam have been established by researchers using genealogical Dna tests. Mitochondrial Eve is estimated to take lived about 200,000 years ago. A paper published in March 2013 determined that, with 95% confidence and that provided in that location are no systematic errors in the study's data, Y-chromosomal Adam lived between 237,000 and 581,000 years ago.[seven] [8]
The MRCA of humans alive today would, therefore, need to have lived more than recently than either.[9] [annotation 3]
Information technology is more complicated to infer human beginnings via autosomal chromosomes. Although an autosomal chromosome contains genes that are passed down from parents to children via independent array from only one of the two parents, genetic recombination (chromosomal crossover) mixes genes from non-sis chromatids from both parents during meiosis, thus changing the genetic limerick of the chromosome.
Time to MRCA estimates [edit]
Different types of MRCAs are estimated to have lived at dissimilar times in the past. These time to MRCA (TMRCA) estimates are besides computed differently depending on the type of MRCA being considered. Patrilineal and matrilineal MRCAs (Mitochondrial Eve and Y-chromosomal Adam) are traced by single gene markers, thus their TMRCA are computed based on Dna test results and established mutation rates equally skilful in genetic genealogy. The time to the genealogical MRCA (near contempo common ancestor by any line of descent) of all living humans cannot exist traced genetically because the Dna of the keen bulk of ancestors is completely lost after a few hundred years. It is therefore computed based on non-genetic, mathematical models and computer simulations.
Since Mitochondrial Eve and Y-chromosomal Adam are traced past single genes via a single ancestral parent line, the time to these genetic MRCAs volition necessarily exist greater than that for the genealogical MRCA. This is because single genes will coalesce more slowly than tracing of conventional man genealogy via both parents. The latter considers only private humans, without taking into account whether whatever cistron from the computed MRCA really survives in every single person in the current population.[eleven]
TMRCA via genetic markers [edit]
Mitochondrial Deoxyribonucleic acid can exist used to trace the ancestry of a set of populations. In this example, populations are defined past the accumulation of mutations on the mtDNA, and special trees are created for the mutations and the order in which they occurred in each population. The tree is formed through the testing of a large number of individuals all over the world for the presence or lack of a certain set of mutations. In one case this is done it is possible to make up one's mind how many mutations carve up i population from another. The number of mutations, together with estimated mutation charge per unit of the mtDNA in the regions tested, allows scientists to make up one's mind the judge fourth dimension to MRCA (TMRCA) which indicates time passed since the populations terminal shared the aforementioned set of mutations or belonged to the aforementioned haplogroup.
In the example of Y-Chromosomal DNA, TMRCA is arrived at in a different way. Y-Dna haplogroups are divers past single-nucleotide polymorphism in various regions of the Y-DNA. The time to MRCA within a haplogroup is divers by the aggregating of mutations in STR sequences of the Y-Chromosome of that haplogroup simply. Y-DNA network assay of Y-STR haplotypes showing a non-star cluster indicates Y-STR variability due to multiple founding individuals. Analysis yielding a star cluster tin be regarded as representing a population descended from a unmarried antecedent. In this case the variability of the Y-STR sequence, also called the microsatellite variation, can be regarded as a measure of the time passed since the ancestor founded this item population. The descendants of Genghis Khan or one of his ancestors represents a famous star cluster that can be dated back to the time of Genghis Khan.[12]
TMRCA calculations are considered critical evidence when attempting to make up one's mind migration dates of diverse populations every bit they spread effectually the world. For example, if a mutation is deemed to take occurred 30,000 years ago, so this mutation should be institute amongst all populations that diverged afterwards this date. If archeological evidence indicates cultural spread and formation of regionally isolated populations so this must exist reflected in the isolation of subsequent genetic mutations in this region. If genetic departure and regional difference coincide it can be concluded that the observed divergence is due to migration every bit evidenced by the archaeological record. However, if the appointment of genetic departure occurs at a unlike time than the archaeological record, then scientists volition have to look at alternate archaeological evidence to explicate the genetic departure. The event is best illustrated in the contend surrounding the demic diffusion versus cultural diffusion during the European Neolithic.[13]
TMRCA of all living humans [edit]
The age of the MRCA of all living humans is unknown. It is necessarily younger than the age of either the matrilinear or the patrilinear MRCA, both of which have an estimated age of betwixt roughly 100,000 and 200,000 years ago.[xiv]
A mathematical, but not-genealogical study by mathematicians Joseph T. Chang, Douglas Rohde and Steve Olson calculated that the MRCA lived remarkably recently, perchance as recently as 300 BCE. This model took into account that people do not truly mate randomly, but that, particularly in the by, people almost always mated with people who lived nearby, and ordinarily with people who lived in their ain town or village. It would have been peculiarly rare to mate with somebody who lived in another state. Withal, Chang et al. found that the rare people who mate with other people far away will in fourth dimension connect the worldwide family tree, and that no population is truly completely isolated.[note iv]
The MRCA of all humans nigh certainly lived in Eastern asia, which would have given them fundamental access to extremely isolated populations in Australia and the Americas. Possible locations for the MRCA include places such equally the Chuckchi and Kamchatka Peninsulas that are close to Alaska, places such as Indonesia and Malaysia that are close to Australia or a identify such every bit Taiwan or Japan that is more intermediate to Australia and the Americas. European colonization of the Americas and Australia was institute by Chang to exist too recent to have had a substantial affect on the age of the MRCA. In fact, if the Americas and Australia had never been discovered by Europeans, the MRCA would only exist about 2.3% further back in the past than information technology is.[17] [18]
Note that the age of the MRCA of a population does not stand for to a population bottleneck, let alone a "offset couple". It rather reflects the presence of a single individual with high reproductive success in the by, whose genetic contribution has become pervasive throughout the population over time. It is too incorrect to assume that the MRCA passed all, or indeed any, genetic information to every living person. Through sexual reproduction, an antecedent passes half of his or her genes to each descendant in the next generation; later more than 32 generations the contribution of a unmarried ancestor would be on the order of two−32, a number proportional to less than a single basepair inside the human genome.[19] [fifteen]
Identical ancestors point [edit]
The MRCA is the about contempo common ancestor shared by all individuals in the population nether consideration. This MRCA may well take contemporaries who are also ancestral to some but not all of the extant population. The identical ancestors point is a point in the past more than remote than the MRCA at which time in that location are no longer organisms which are ancestral to some but not all of the modern population. Due to pedigree collapse, modern individuals may all the same exhibit clustering, due to vastly different contributions from each of ancestral population.[15]
Come across besides [edit]
- Cladistics
- Mutual descent
- Coalescent theory, a retrospective model of population genetics
- Genealogy, the study of families and the tracing of their lineages and history
- Genetic distance, the genetic deviation between species or between populations within a species
- Everyman common ancestor, an analogous concept in graph theory and computer scientific discipline
- Phylogenetic tree, a branching diagram or "tree" showing the inferred evolutionary relationships among various biological species
- Timeline of evolution, outlines the major events in the development of life on the planet Earth
- Timeline of homo evolution, outlines the major events in the development of the human species
Notes [edit]
- ^ MRCA is at present more frequently used for the common ancestor of subgroups within a species, and LCA for the mutual ancestor of two separate species.[ citation needed ] The term "concestor" (coined by Nicky Warren) is used by Richard Dawkins in The Ancestor'southward Tale (2004).
- ^ The limerick of the LUCA is not directly accessible as a fossil, but can be studied by comparing the genomes of its descendants, organisms living today. By this means, a 2016 study identified a set of 355 genes inferred to have been present in the LUCA.[4]
- ^ Notions such as Mitochondrial Eve and Y-chromosomal Adam yield common ancestors that are more aboriginal than for all living humans.[10]
- ^ Rohde[15] calculates an age of 3,000 to 7,000 years based on a non-genetic, mathematical model that assumes random mating, although information technology has taken into business relationship important aspects of human population substructure such as assortative mating and historical geographical constraints on interbreeding. This range is consistent with the age of 3,100 years calculated for the MRCA of the JC virus, a ubiquitous human polyomavirus unremarkably transmitted from parents to children.[xvi]
References [edit]
- ^ Brower AVZ, Schuh RT (2021). Biological Systematics: Principles and Applications (tertiary edn.). Ithaca, NY: Cornell Academy Press.
- ^ Doolittle WF (February 2000). "Uprooting the tree of life". Scientific American. 282 (two): xc–95. Bibcode:2000SciAm.282b..90D. doi:ten.1038/scientificamerican0200-90. PMID 10710791.
- ^ Glansdorff N, Xu Y, Labedan B (2008). "The last universal common ancestor: emergence, constitution and genetic legacy of an elusive precursor". Biology Directly. 3: 29. doi:ten.1186/1745-6150-3-29. PMC2478661. PMID 18613974.
- ^ Wade, Nicholas (25 July 2016). "Meet Luca, the Antecedent of All Living Things". New York Times . Retrieved 25 July 2016.
- ^ Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P; Doerks; von Mering; Creevey; Snel; Bork (2006). "Toward automatic reconstruction of a highly resolved tree of life". Scientific discipline. 311 (5765): 1283–87. Bibcode:2006Sci...311.1283C. CiteSeerXx.i.1.381.9514. doi:x.1126/science.1123061. PMID 16513982. S2CID 1615592.
{{cite journal}}: CS1 maint: multiple names: authors listing (link) - ^ Eizirik, Eastward.; Murphy, W.J.; Koepfli, K.P.; Johnson, West.East.; Dragoo, J.W.; O'Brien, S.J. (2010). "Pattern and timing of the diversification of the mammalian order Carnivora inferred from multiple nuclear gene sequences". Molecular Phylogenetics and Development. 56 (ane): 49–63. doi:10.1016/j.ympev.2010.01.033. PMC7034395. PMID 20138220.
- ^ Mendez, Fernando; Krahn, Thomas; Schrack, Bonnie; Krahn, Astrid-Maria; Veeramah, Krishna; Woerner, August; Fomine, Forka Leypey Mathew; Bradman, Neil; Thomas, Mark; Karafet, Tatiana 1000.; Hammer, Michael F. (7 March 2013). "An African American paternal lineage adds an extremely ancient root to the human Y chromosome phylogenetic tree" (PDF). American Journal of Human being Genetics. 92 (3): 454–59. doi:10.1016/j.ajhg.2013.02.002. PMC3591855. PMID 23453668. (primary source)
- ^ Barrass, Colin (6 March 2013). "The father of all men is 340,000 years old". New Scientist. Retrieved 13 March 2013.
- ^ Dawkins, Richard (2004). The Ancestor'due south Tale, A Pilgrimage to the Dawn of Life. Boston: Houghton Mifflin Company. ISBN978-0-618-00583-three.
- ^ Hartwell 2004, p. 539.
- ^ Chang, Joseph T.; Donnelly, Peter; Wiuf, Carsten; Hein, Jotun; Slatkin, Montgomery; Ewens, Westward. J.; Kingman, J. F. C. (1999). "Recent mutual ancestors of all present-day individuals" (PDF). Advances in Applied Probability. 31 (4): 1002–26, word and author's reply, 1027–38. CiteSeerX10.1.1.408.8868. doi:10.1239/aap/1029955256. S2CID 1090239. Retrieved 2008-01-29 .
- ^ Tatiana Zerjal (2003), The Genetic Legacy of the Mongols, "Archived re-create" (PDF). Archived from the original (PDF) on 2012-07-10. Retrieved 2012-06-28 .
{{cite spider web}}: CS1 maint: archived copy as title (link) - ^ Morelli 50, Contu D, Santoni F, Whalen MB, Francalacci P; Contu; Santoni; Whalen; Francalacci; Cucca; et al. (2010). Lalueza-Play a joke on, Carles (ed.). "A Comparing of Y-Chromosome Variation in Sardinia and Anatolia Is More Consistent with Cultural Rather than Demic Diffusion of Agriculture". PLOS ONE. 5 (4): e10419. Bibcode:2010PLoSO...510419M. doi:10.1371/periodical.pone.0010419. PMC2861676. PMID 20454687.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Poznik, GD; Henn, BM; Yee, MC; Sliwerska, E; Euskirchen, GM; Lin, AA; Snyder, M; Quintana-Murci, L; Kidd, JM; Underhill, PA; Bustamante, CD (2013). "Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females". Science. 341 (6145): 562–65. Bibcode:2013Sci...341..562P. doi:10.1126/science.1237619. PMC4032117. PMID 23908239.
- ^ a b c Rohde DL, Olson S, Chang JT; Olson; Chang (September 2004). "Modelling the recent common ancestry of all living humans" (PDF). Nature. 431 (7008): 562–66. Bibcode:2004Natur.431..562R. CiteSeerXx.ane.1.78.8467. doi:10.1038/nature02842. PMID 15457259. S2CID 3563900.
{{cite periodical}}: CS1 maint: multiple names: authors list (link) - ^ Shackelton, L. A.; Rambault, A; Pybus, O; Holmes, Eastward (2006). "JC Virus Development and Its Association with Human Populations". Journal of Virology. 80 (twenty): 9928–9933. doi:x.1128/JVI.00441-06. PMC1617318. PMID 17005670.
- ^ "Roots of Human Family Tree Are Shallow". July 2006.
- ^ "Archived copy" (PDF). Archived from the original (PDF) on 2018-12-30. Retrieved 2018-05-01 .
{{cite spider web}}: CS1 maint: archived copy as championship (link) - ^ Zhaxybayeva, Olga; Lapierre, Pascal; Gogarten, J. Peter (May 2004). "Genome mosaicism and organismal lineages" (PDF). Trends in Genetics. 20 (v): 254–60. CiteSeerX10.one.ane.530.7843. doi:10.1016/j.tig.2004.03.009. PMID 15109780. Retrieved 2009-02-19 .
The Transport of Theseus paradox […] is often invoked to illustrate this point […]. Even moderate levels of gene transfer volition brand information technology incommunicable to reconstruct the genomes of early ancestors; …
Further reading [edit]
- Hartwell, Leland (2004). Genetics: From Genes to Genomes (2nd ed.). Maidenhead: McGraw-Hill. ISBN978-0-07-291930-i.
- Walsh B (June 2001). "Estimating the time to the most contempo common antecedent for the Y chromosome or mitochondrial DNA for a pair of individuals" (PDF). Genetics. 158 (2): 897–912. doi:10.1093/genetics/158.2.897. PMC1461668. PMID 11404350.
Source: https://en.wikipedia.org/wiki/Most_recent_common_ancestor
Posted by: gonzalezesifer88.blogspot.com

0 Response to "What Is The Most Recent Common Ancestor Of Fungi And Animals Quizlet"
Post a Comment